High throughput RNA sequencing (RNA-seq) has unveiled unexpectedly complex transcriptional outputs in bacteria, adding large and heterogeneous inventories of small non-protein coding transcripts (sRNAs) to the translation-related ribosomal (rRNA), transfer (tRNA) and messenger (mRNA) RNAs. Most sRNAs act as post-transcriptional regulators of gene expression, governing extensive regulatory networks that help bacteria cope with and adapt to changing environments. However, sRNAs are systematically overlooked in the annotation of bacterial genomes and riboregulation remains understudied in most environmentally relevant non-classical model microbes.

Regulation by RNA in Rhizobia
In the RhizoRNA Lab we study RNA regulation in a group of α-proteobacteria, known as rhizobia, that establish nitrogen-fixing endosymbioses with legume plants. These mutualistic associations render plant growth independent of environmentally unfriendly chemical fertilizers and are, therefore, pillars of planet sustainability. Besides, rhizobia-legume symbioses provide invaluable experimental systems for deciphering the adaptive traits operating in bacteria during chronic infection of eukaryotic hosts. Rhizobia persist in soil under usually harsh conditions, actively compete in the rhizosphere with beneficial and pathogenic microorganisms, and finally induce the host plant to form root nodules that accommodate nitrogen-fixing differentiated bacteroids intracellularly. During this symbiotic transition rhizobial gene expression undergoes continuous and profound reprograming in response to abiotic stimuli and plant-derived cues. We are interested in unravelling the roles of sRNAs in these adaptive regulatory networks and their impact in rhizobial physiology, metabolism and symbiotic nitrogen-fixation. Given that RNA regulation relies mostly on modifiable base-pairing interactions, our longer-term goal is to exploit the functional plasticity of sRNAs for the engineering of the legumes rhizobiome and symbiotic efficiency of rhizobia in the sustainable agricultural practice.
Research Lines
1. Structure of the rhizobial non-coding transcriptome.
We use diverse RNA-seq protocols to update the annotation of rhizobial genomes with the inclusion of trans-acting sRNAs, antisense sRNAs (asRNAs) and untranslated regions (5’/3’-UTRs) of mRNAs. These new genome annotations are used as a reference to identify differentially expressed sRNAs in symbiotically relevant conditions by classical RNA-seq protocols.
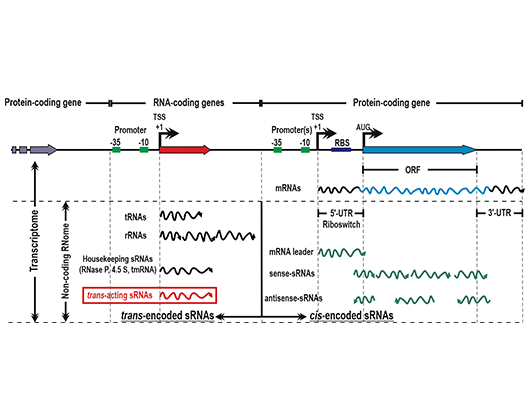
The complexity of the prokaryotic transcriptome as revealed by strand-specific RNA-seq. Categories of RNA species according to the location of their coding loci with respect to the annotated protein-coding genes. Number of sRNAs of each class expressed by S. meliloti are indicated. Most sRNAs are uncharacterized regulators of gene expression.
2. Riboregulation of rhizobial metabolism.
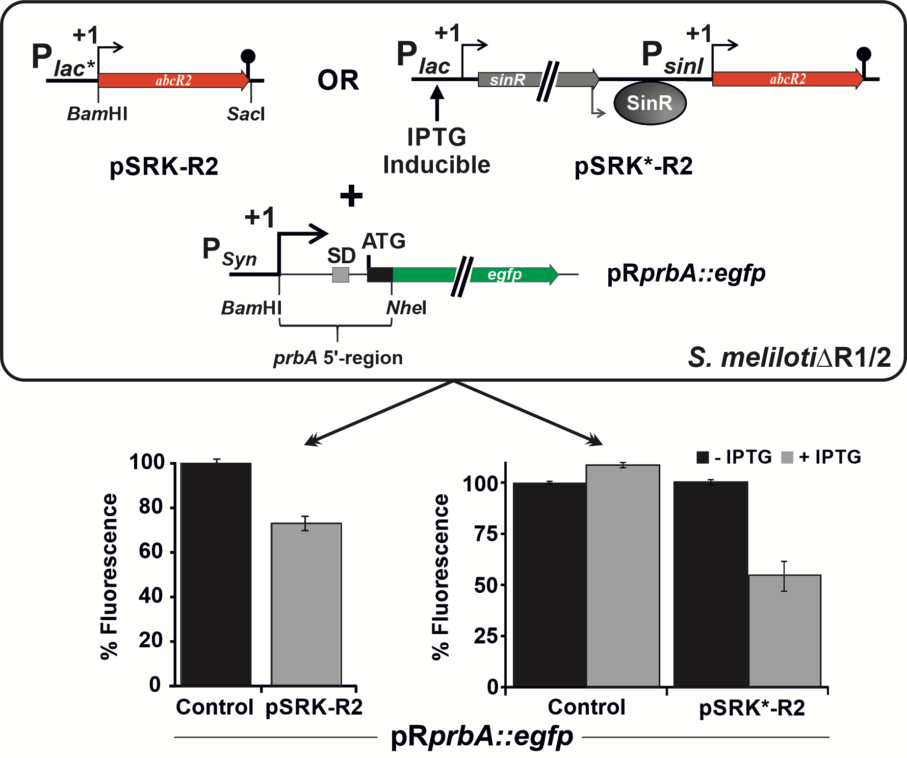
We are currently investigating in detail the function and activity mechanisms of three sRNAs called AbcR1, AbcR1 and NfeR1 that pervasively regulate mRNAs devoted to nutrient uptake and metabolism in the alfalfa symbiont Sinorhizobium meliloti. These sRNAs belong to the most studied class of riboregulators in bacteria, the so-called trans-sRNAs, which typically regulate translation and stability of multiple mRNAs by protein-assisted short and discontinuous base-pairing at the 5’-region of the target transcripts.
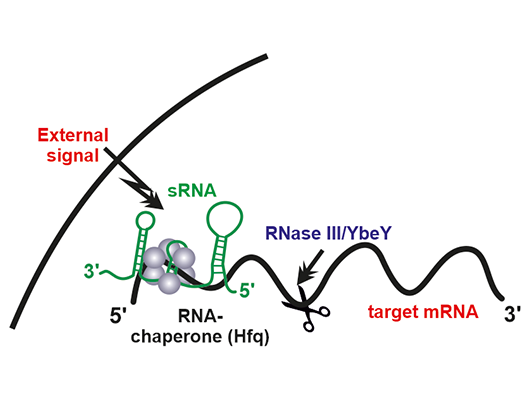
Activity mechanism of the bacterial trans-sRNAs. These sRNAs are differentially transcribed from independent promoters within intergenic regions in response to specific external signals and base-pair to the translation initiation region of multiple trans-encoded target mRNAs. These antisense interactions are usually assisted by and RNA chaperone such as Hfq, which acts as matchmaker. sRNA-mRNA interaction results in blocking of translation and/or promotes mRNA degradation by RNases (e.g. YbeY and RNaseIII).
3. Regulation of symbiotic genes by antisense transcription.
Transcription from the antisense strand of protein-coding genes is pervasive in S. meliloti, being particularly abundant in genuine symbiotic regions of the pSymA megaplasmid. We study how asRNAs affect expression of nodulation and nitrogen-fixation genes in S. meliloti.
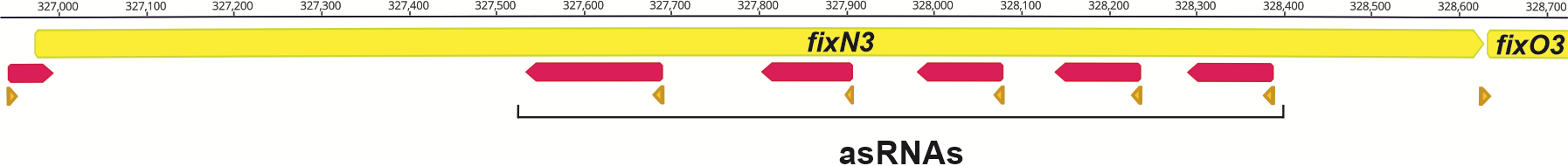
Antisense transcription of the S. meliloti fixN3 gene.
4. RNA-binding proteins and ribonucleases.
Using S. meliloti as model experimental system we investigate the function in riboregulation of the widespread bacterial chaperone Hfq and the endoribonucleases YbeY and RNase III. YbeY is a remarkable RNase identified in our laboratory that has an unprecedented ability among bacterial endoribonucleases to cleave both single- and double-stranded RNA substrates. We are also interested in the identification of novel RNA chaperones and the characterization of the whole set of ribonucleases in this bacterium.
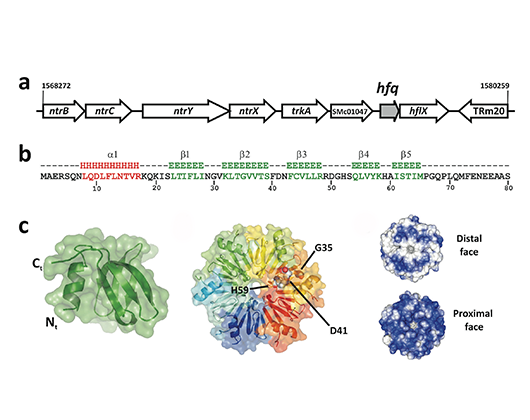
The S. meliloti Hfq protein. Genomic region, amino acid sequence and predicted structure.
Methodological approaches
- Accurate identification of 5’ and 3’ boundaries of all cellular transcripts and primary characterization of the rhizobial transcriptomes: cutting-edge RNA-seq protocols (dRNA-seq, Cappable-seq, tagRNA-seq, or Term-seq).
- Functional characterization of sRNAs: molecular genetics, biochemistry, microbial/plant physiology, microscopy, bioinformatics and genomics.
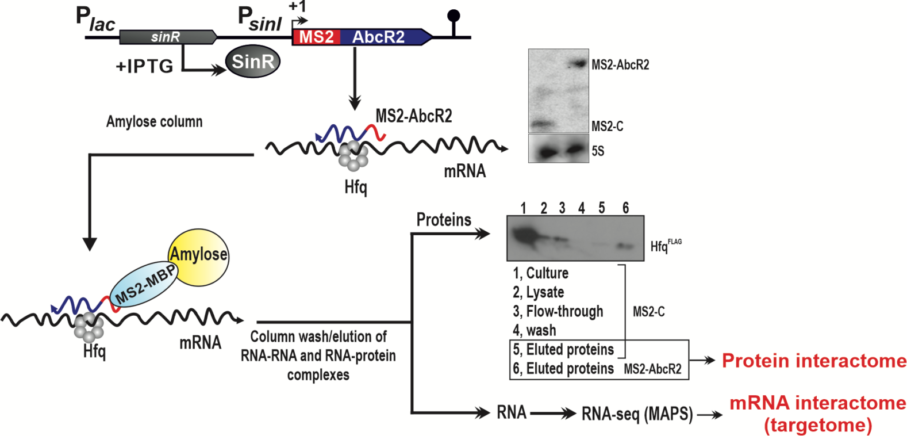
- Genome-wide identification of RNA-RNA, protein-RNA and protein-DNA interactions: methods based on high-throughput sequencing such as MAPS (MS2-affinity purification coupled with RNA sequencing), CLASH-seq (UV-Crosslinking, Ligation And Sequencing of RNA-RNA Hybrids), or ChIP-seq (chromatin immunoprecipitation).
- Catalytic versatility of the YbeY RNase: in vitro assays, crystallography, and structural biology.